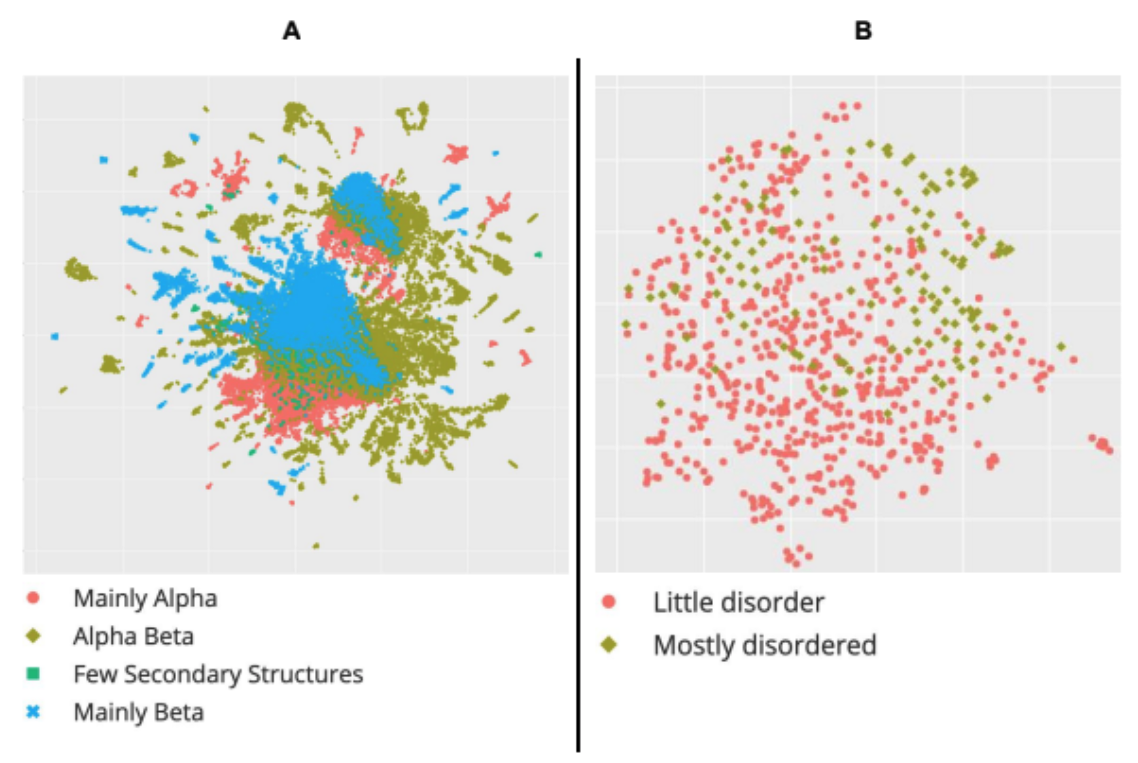
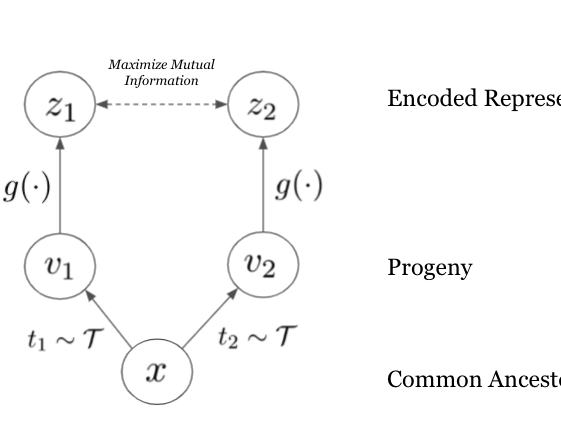
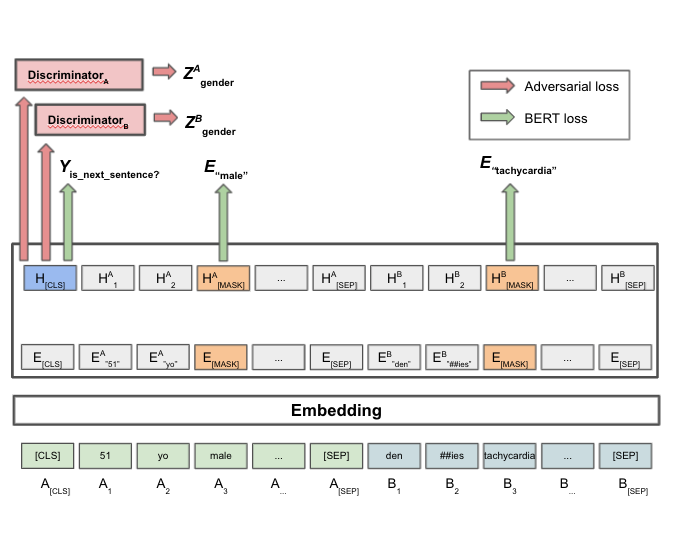
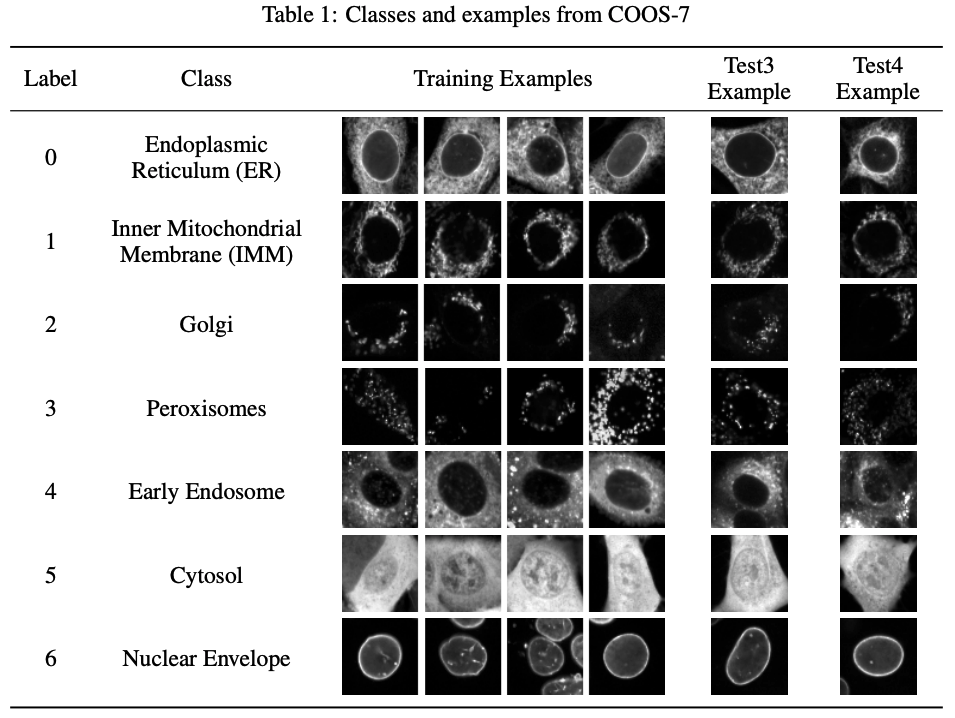
Genome modeling and design across all domains of life with Evo 2
Garyk Brixi
,
Matthew G. Durrant
,
Jerome Ku
,
Michael Poli
,
Greg Brockman
,
Daniel Chang
,
Gabriel A. Gonzalez
,
Samuel H. King
,
David B. Li
,
Aditi T. Merchant
,
Mohsen Naghipourfar
,
Eric Nguyen
,
Chiara Ricci-Tam
,
David W. Romero
,
Gwanggyu Sun
,
Ali Taghibakshi
,
Anton Vorontsov
,
Brandon Yang
,
Myra Deng
,
Liv Gorton
,
Nam Nguyen
,
Nicholas K. Wang
,
Etowah Adams
,
Stephen A. Baccus
,
Steven Dillmann
,
Stefano Ermon
,
Daniel Guo
,
Rajesh Ilango
,
Ken Janik
,
Amy X. Lu
,
Reshma Mehta
,
Mohammad R.K. Mofrad
,
Madelena Y. Ng
,
Jaspreet Pannu
,
Christopher Ré
,
Jonathan C. Schmok
,
John St. John
,
Jeremy Sullivan
,
Kevin Zhu
,
Greg Zynda
,
Daniel Balsam
,
Patrick Collison
,
Anthony B. Costa
,
Tina Hernandez-Boussard
,
Eric Ho
,
Ming-Yu Liu
,
Thomas McGrath
,
Kimberly Powell
,
Dave P. Burke
,
Hani Goodarzi
,
Patrick D. Hsu
,
Brian L. Hie
bioRxiv, 2025
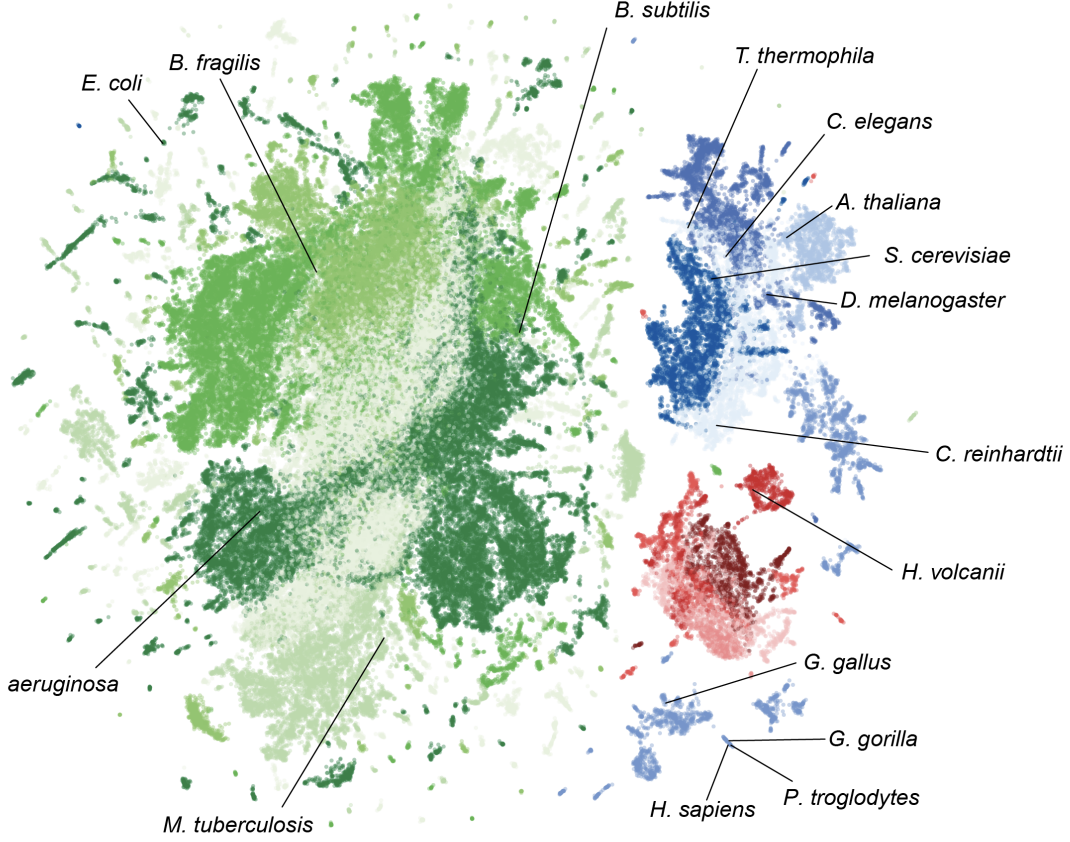
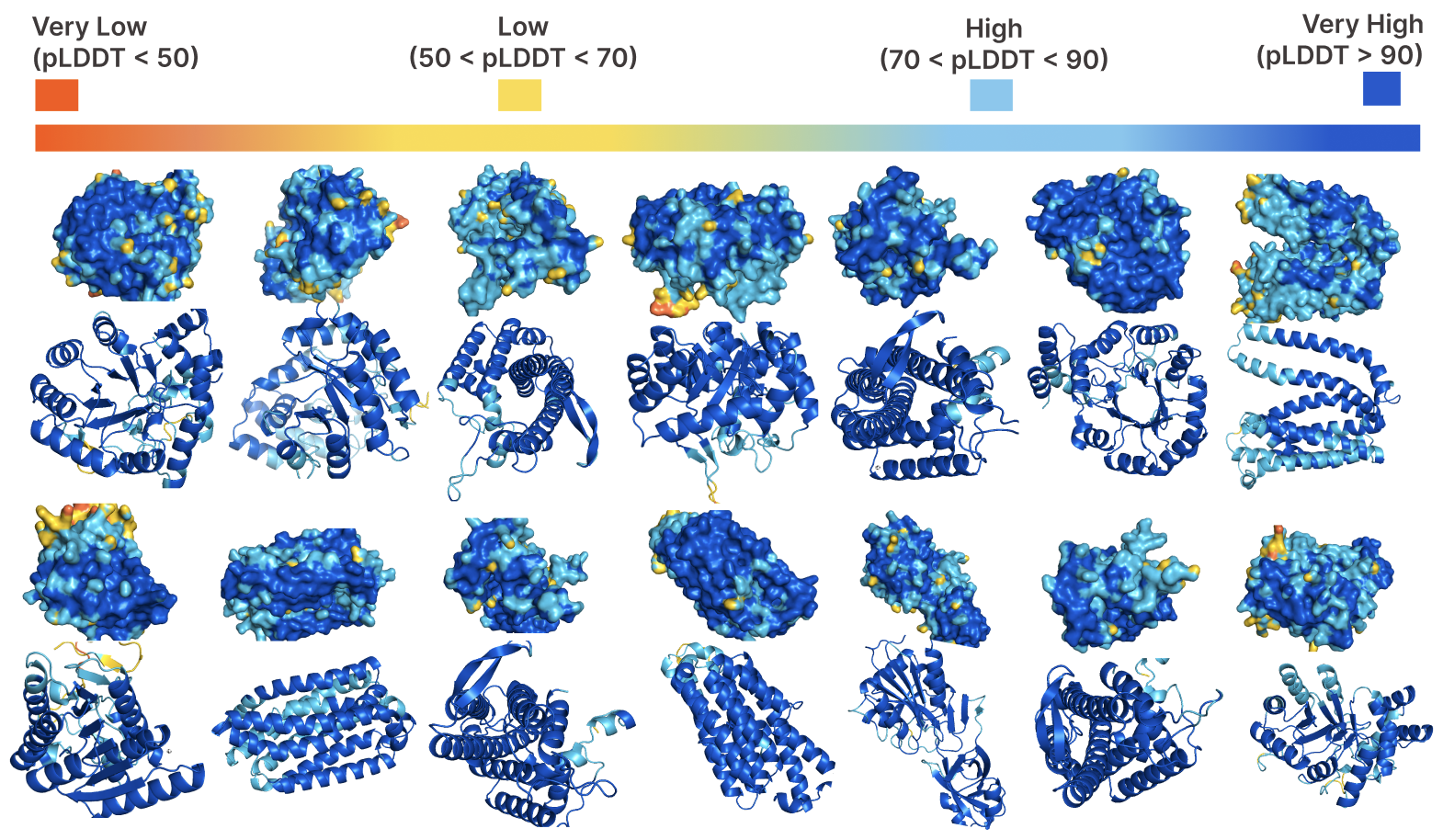
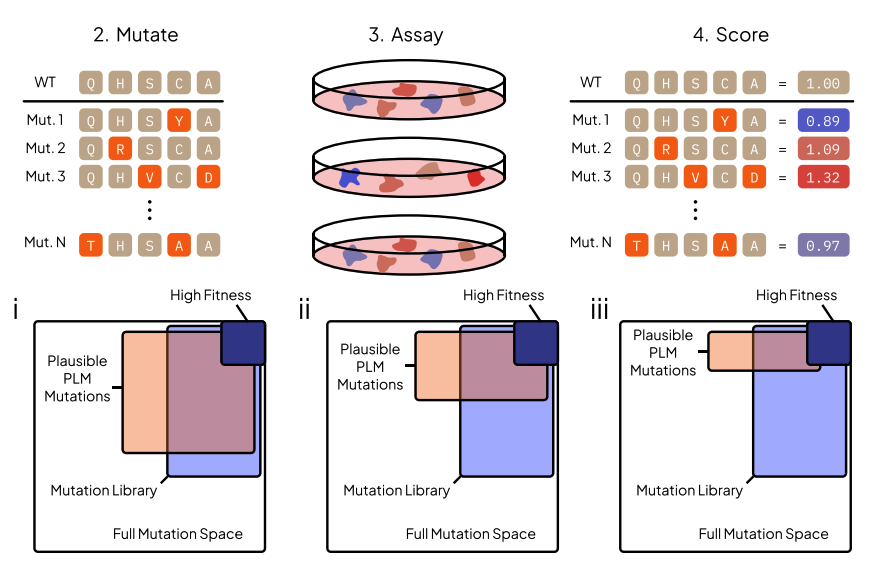
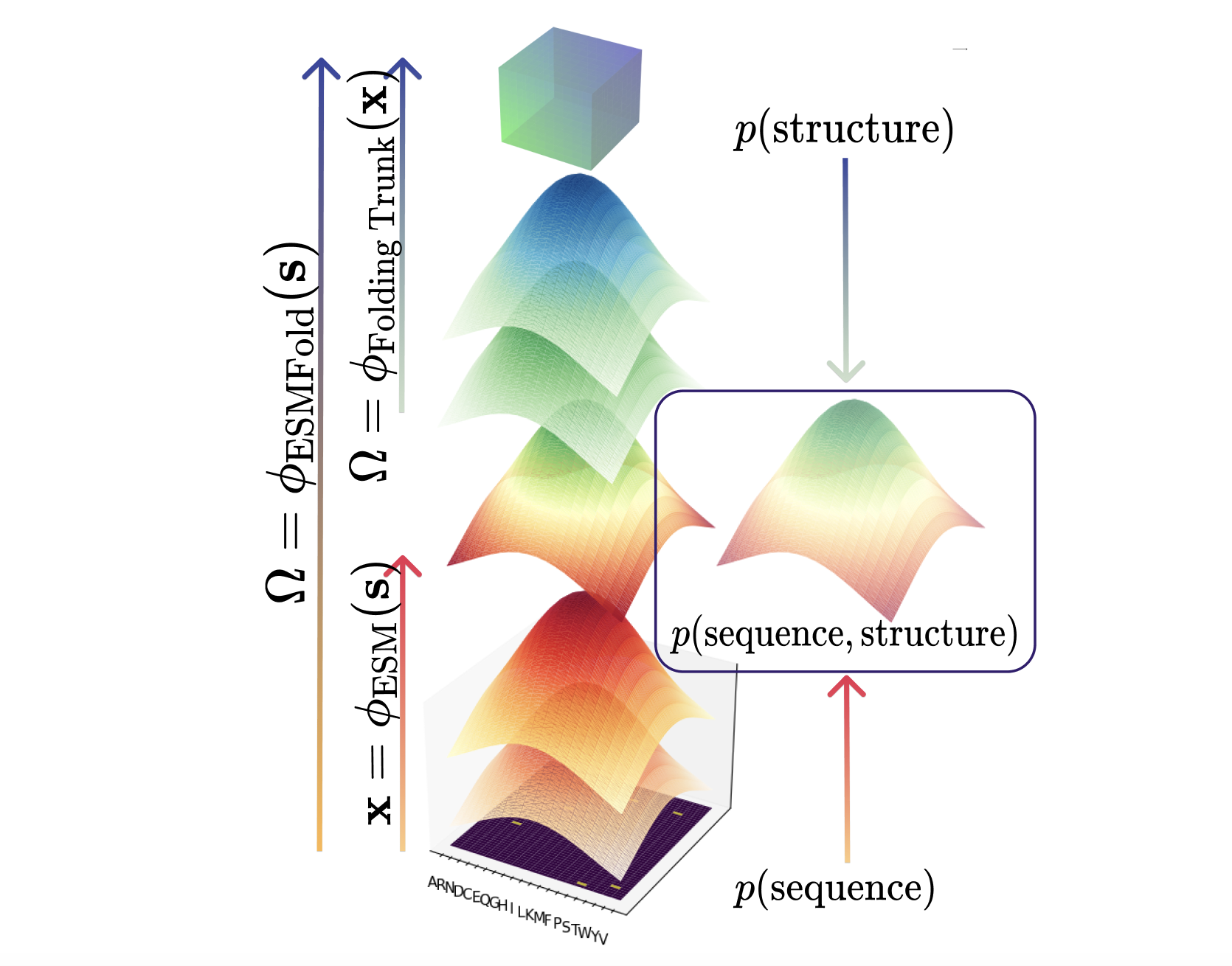
Evo 2 is a 40B parameter genomic foundation model capable of predicting functional impacts of genetic variations, autonomously learning biological features, and generating novel genomic sequences across all domains of life.